Using Estimator1D¶
The nmrespy.Estimator1D class is provided for the consideration of 1D NMR data.
Generating an instance¶
There are a few ways to create a new instance of the estimator depending on the source of the data.
Bruker data¶
It is possible to load both raw FID data and processed spectral data from
Bruker using new_bruker(). All that is needed is
the path to the dataset:
If you wish to import an FID, set the path as
"<path_to_data>/<expno>/". There should be anfidfile and anacqusfile directly under this directory. The data in thefidfile will be imported, and the artefact from digital filtering will be removed by a first-order phase shift.Note
If you import FID data, there is a high chance that you will need to phase the data, and apply baseline correction before proceeding to run estimation. Look at
phase_data()andbaseline_correction(), respectively.>>> import nmrespy as ne >>> estimator = ne.Estimator1D.new_bruker("/home/simon/nmr_data/andrographolide/1") >>> estimator.phase_data(p0=2.653, p1=-5.686, pivot=13596) >>> estimator.baseline_correction()
To import processed data, set the path as
"<path_to_data>/<expno>/pdata/<procno>". There should be a1rfile and aprocsfile directly under this directory. The data in1rwill be Inverse Fourier Transformed, and the resulting time-domain signal is sliced so that only the first half is retained.Note
It can be more convienient to provide processed data, even though the data will be converted to the time-domain for estimation, as you can then rely on TopSpin’s automated processing scripts to phase and baseline correct. However, you should not apply any window function to the data other than exponential line broadening.
>>> import nmrespy as ne >>> estimator = ne.Estimator1D.new_bruker("home/simon/nmr_data/andrographolide/1/pdata/1") >>> # Note there is no need for extra data-processing steps
Simulated data from a set of signal parameters¶
You can create an estimator with synthetic data constructed from known
parameters using new_from_parameters().
The parameters must be provided as a 2D NumPy array with params.shape[1] ==
4. Each row should contain a signal’s amplitude, phase (rad), frequency
(Hz), and damping factor (s⁻¹).
>>> import nmrespy as ne
>>> import numpy as np
>>> # Using frequencies of 2,3-Dibromopropanoic acid @ 500MHz
>>> params = np.array([
... [1., 0., 1864.4, 7.],
... [1., 0., 1855.8, 7.],
... [1., 0., 1844.2, 7.],
... [1., 0., 1835.6, 7.],
... [1., 0., 1981.4, 7.],
... [1., 0., 1961.2, 7.],
... [1., 0., 1958.8, 7.],
... [1., 0., 1938.6, 7.],
... [1., 0., 2265.6, 7.],
... [1., 0., 2257.0, 7.],
... [1., 0., 2243.0, 7.],
... [1., 0., 2234.4, 7.],
... ])
>>> sfo = 500.
>>> estimator = ne.Estimator1D.new_from_parameters(
... params=params,
... pts=2048, # FID made with 2048 points
... sw=1.2 * sfo, # sweep width set to 1.2 ppm
... offset=4.1 * sfo, # transmitter offset set to 4.1 ppm
... sfo=sfo, # transmitter frequecy set to 500 MHz
... snr=40., # signal-to-noise ratio of the FID set to 40 dB
... )
Note
For the rest of this section, we will be using the estimator created in the above code snippet.
Simulated data from Spinach¶
Assuming you have installed the relevant requirements,
you can create an estimator instance with data simulated using Spinach with
new_spinach(). The spin system is defined by a
specification of isotropic chemical shifts and scalar couplings:
For the chemical shifts, a list of floats is required.
For J-couplings, a list with 3-element tuples of the form
(spin1, spin2, coupling)is required. N.B. the spin indices start at ``1`` rather than ``0``.
It can take some time to run this function is it involves (a) starting up MATLAB and (b) running a simulation of the experiment.
>>> import nmrespy as ne
>>> # 2,3-Dibromopropanoic acid
>>> shifts = [3.7, 3.92, 4.5]
>>> couplings = [(1, 2, -10.1), (1, 3, 4.3), (2, 3, 11.3)]
>>> sfo = 500.
>>> estimator = ne.Estimator1D.new_spinach(
... shifts=shifts,
... couplings=couplings,
... pts=2048,
... sw=1.2 * sfo,
... offset=4.1 * sfo,
... sfo=sfo,
... )
Viewing and accessing the dataset¶
You can inspect the data associated with the estimator with
view_data(), which loads an interactive
matplotlib figure:
>>> estimator.view_data(freq_unit="ppm")
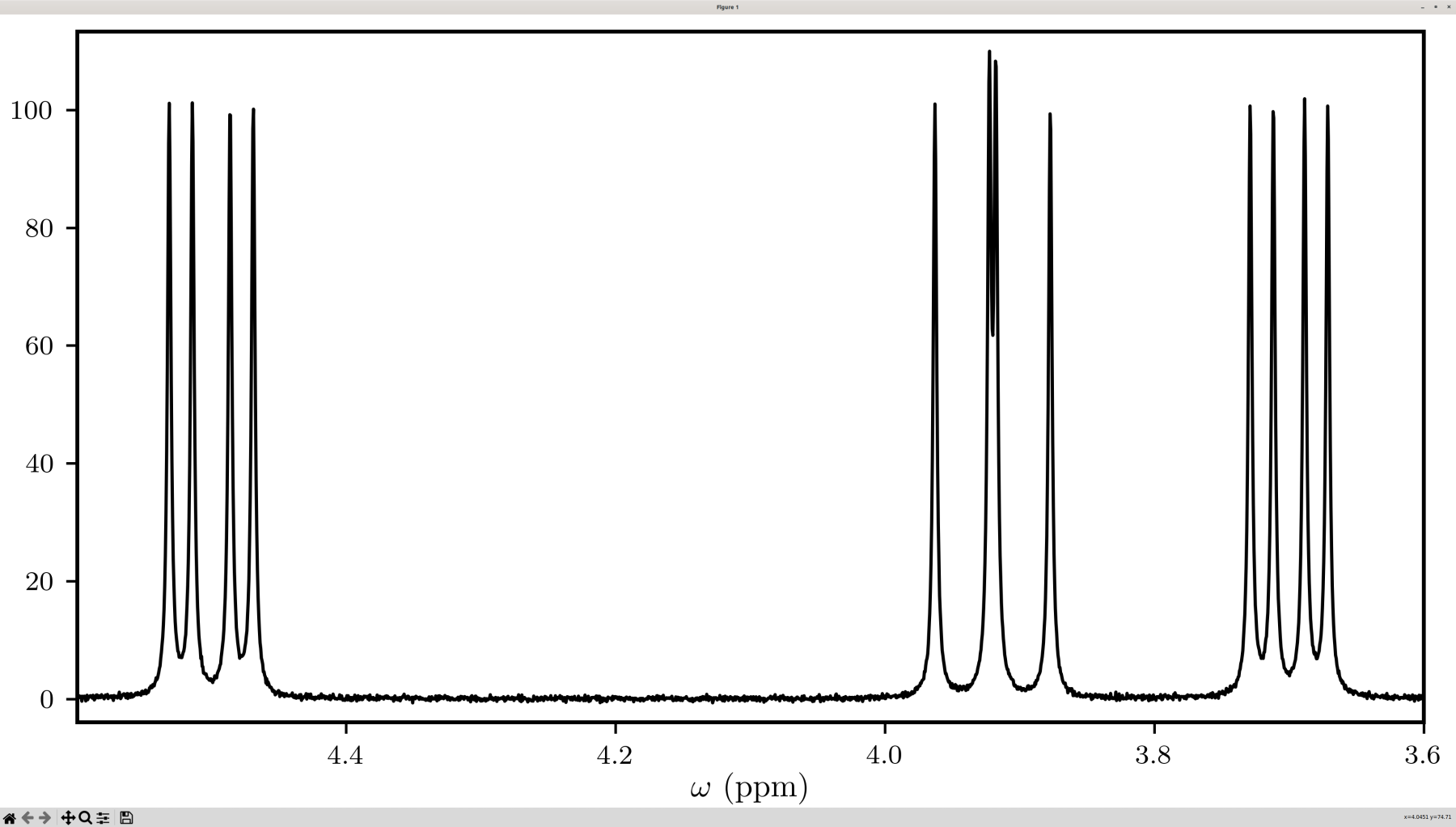
You can access the time-domain data with the
data() property,
and the associated time-points can be retrieved using
get_timepoints(). The spectral data is accessed
with spectrum(), and the corresponding chemical
shifts with get_shifts().
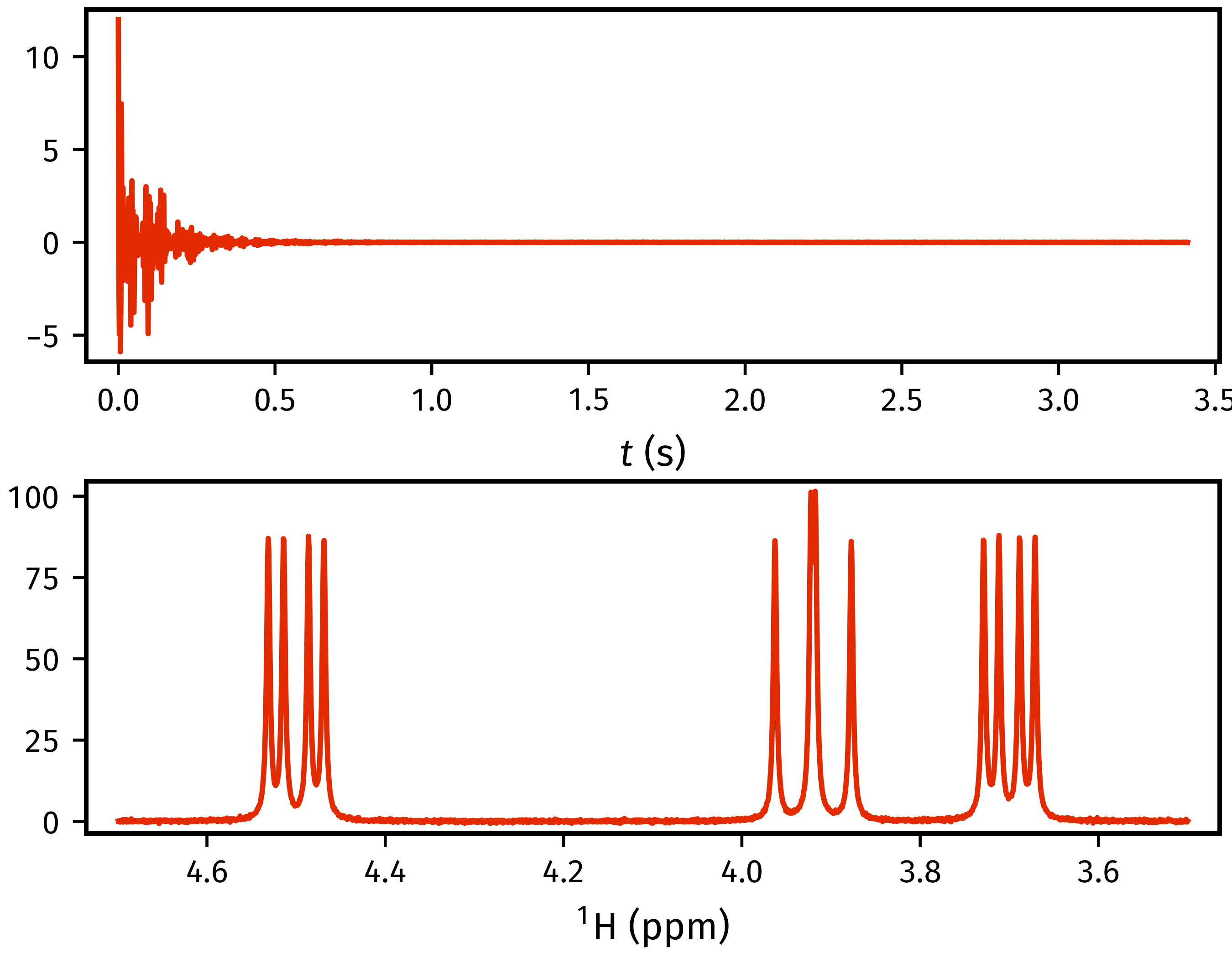
>>> import matplotlib.pyplot as plt
>>> fid = estimator.data
>>> tp = estimator.get_timepoints()[0]
>>> spectrum = estimator.spectrum
>>> shifts = estimator.get_shifts(unit="ppm")[0]
>>> fig, axs = plt.subplots(nrows=2)
>>> axs[0].plot(tp, fid.real)
[<matplotlib.lines.Line2D object at 0x7f2c0e0aa680>]
>>> axs[0].set_xlabel("$t$ (s)")
Text(0.5, 0, '$t$ (s)')
>>> axs[1].plot(shifts, spectrum.real)
[<matplotlib.lines.Line2D object at 0x7f2c0e0aa8c0>]
>>> # Flip x-axis limits (ensure plotting from high to low shifts)
>>> axs[1].set_xlim(reversed(axs[1].get_xlim()))
(4.759384765624999, 3.4400292968749997)
>>> axs[1].set_xlabel("$^1$H (ppm)")
Text(0.5, 0, '$^1$H (ppm)')
>>> # The exact appearence of the generated figure may slightly differ to
>>> # the one below; that'll just be due to customisations I have made to mpl.
>>> plt.show()
Estimating the dataset¶
The generation of parameter estimates is facilitated using the
estimate() method. In most scenarios,
it will not be computationally feasible to estimate the entire FID at once, due
to the number of constituent datapoints and signals. For this reason, NMR-EsPy generates
frequency-filtered “sub-FIDs” to break the problem down into more manageable
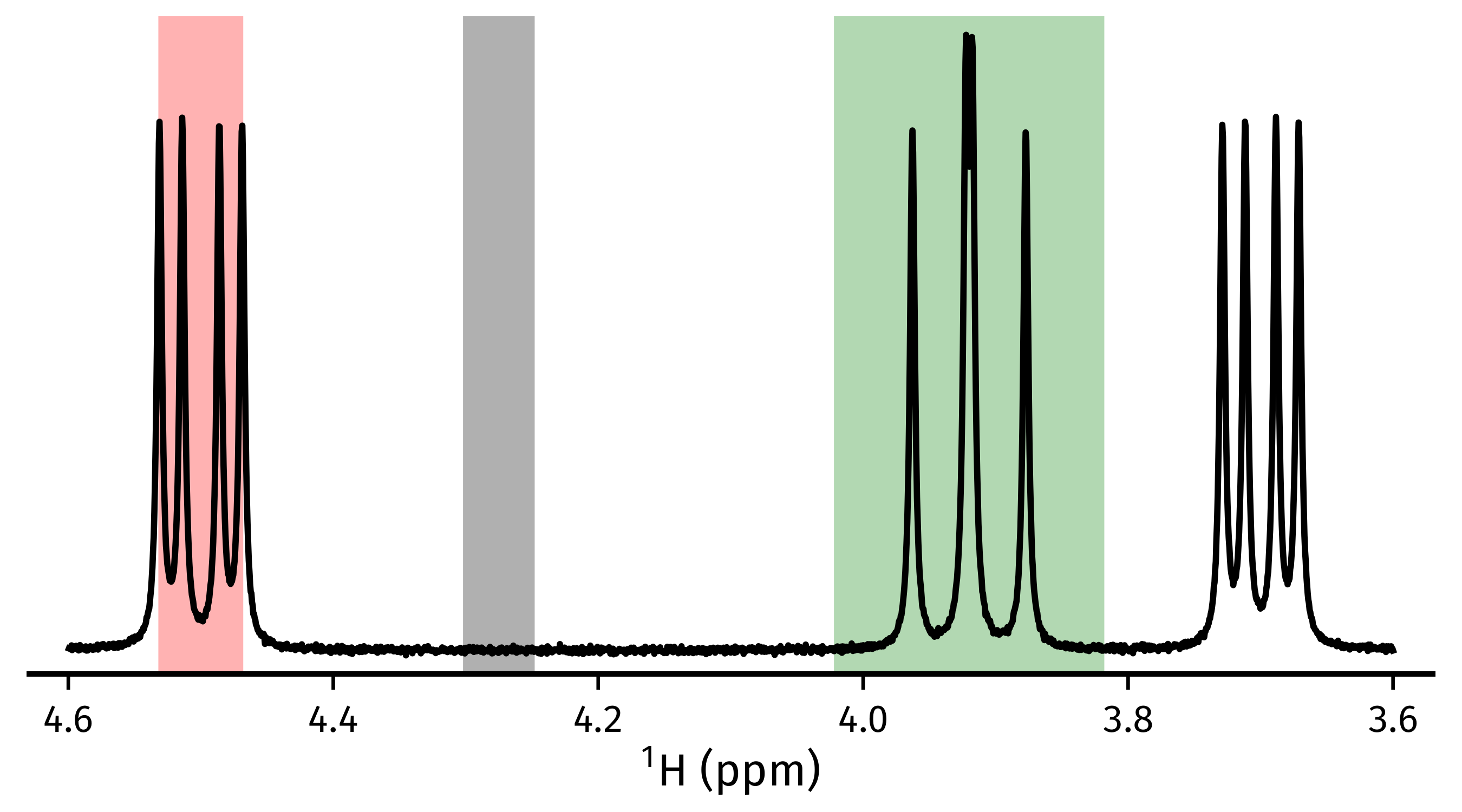
chunks. To create suitable sub-FIDs, it is important to select regions in which
all signals of interest fully reside within its bounds. As well as this, a
region that is devoid of signals (the “noise region”) must be indicated. In the
figure below, the red region would be inappropriate as the signals clearly do
not reside within it fully. The green region is acceptable, as the signals do
abide by this. Finally, the grey region is a suitable noise region as it only
comprises points in the baseline.
For our dataset, we will estimate three regions, encompassing each multiplet
structure in the spectrum. Each region must be given as a tuple of 2 floats,
specifying the left and right boundaries of the region of interest (the order
of these doesn’t matter). By default, these are assumed to be given in Hz,
unless region_unit is set to "ppm".
>>> regions = [(4.6, 4.4), (4.02, 3.82), (3.8, 3.6)]
>>> noise_region = (4.3, 4.25)
>>> for region in regions:
... estimator.estimate(
... region=region, noise_region=noise_region, region_unit="ppm",
... )
...
┌─────────────────────────────────────┐
│ESTIMATING REGION: 4.6 - 4.4 ppm (F1)│
└─────────────────────────────────────┘
┌───────────┐
│MPM STARTED│
└───────────┘
--> Pencil Parameter: 125
--> Hankel data matrix constructed:
Size: 251 x 126
Memory: 0.4826MiB
--> Performing Singular Value Decomposition...
--> Computing number of oscillators...
Number of oscillators will be estimated using MDL
Number of oscillations: 4
--> Computing signal poles...
--> Computing complex amplitudes...
--> Checking for oscillators with negative damping...
None found
┌────────────┐
│MPM COMPLETE│
└────────────┘
Time elapsed: 0 mins, 0 secs, 45 msecs
┌────────────────────┐
│OPTIMISATION STARTED│
└────────────────────┘
┌───────────────────────────┐
│TRUST NCG ALGORITHM STARTED│
└───────────────────────────┘
┌───────┬──────────────┬──────────────┬──────────────┐
│ Iter. │ Objective │ Grad. Norm │ Trust Radius │
├───────┼──────────────┼──────────────┼──────────────┤
│ 0 │ 0.0106245 │ 0.0653396 │ 1 │
│ 6 │ 0.00529026 │ 4.06591e-09 │ 1 │
└───────┴──────────────┴──────────────┴──────────────┘
Optimiser successfully converged.
┌────────────────────────────┐
│TRUST NCG ALGORITHM COMPLETE│
└────────────────────────────┘
Time elapsed: 0 mins, 0 secs, 34 msecs
┌─────────────────────┐
│OPTIMISATION COMPLETE│
└─────────────────────┘
Time elapsed: 0 mins, 0 secs, 101 msecs
┌───────────────────────────────────────┐
│ESTIMATING REGION: 4.02 - 3.82 ppm (F1)│
└───────────────────────────────────────┘
--snip--
┌─────────────────────────────────────┐
│ESTIMATING REGION: 3.8 - 3.6 ppm (F1)│
└─────────────────────────────────────┘
--snip--
Inspecting estimation results¶
Note
Result indices
Each time the estimate() method is called, the
result is appended to a list containing all the generated results. For many
methods that make use of estimation results, an argument called indices
exists. This lets you specify the results you are interested in. By default
(indices = None) all results will be used. A subset of the results can
be considered by including a
list of integers. For example indices = [0, 2] would mean only the 1st
and 3rd results acquired with the estimator are considered.
A NumPy array of the generated results can be acquired using
get_params(). The corresponding errors associated
with the signal parameters are obtained with get_errors().
>>> # All params, frequencies in Hz:
>>> estimator.get_params()
[[ 1.0018e+00 1.5921e-03 1.8356e+03 7.0187e+00]
[ 1.0003e+00 2.4881e-03 1.8442e+03 6.9968e+00]
[ 1.0024e+00 1.5817e-03 1.8558e+03 7.0281e+00]
[ 1.0008e+00 9.1591e-04 1.8644e+03 7.0007e+00]
[ 1.0022e+00 7.1936e-04 1.9386e+03 7.0109e+00]
[ 9.9470e-01 -7.4609e-04 1.9588e+03 6.9866e+00]
[ 1.0080e+00 -1.0112e-03 1.9612e+03 7.0448e+00]
[ 1.0009e+00 -7.1398e-04 1.9814e+03 7.0131e+00]
[ 1.0003e+00 1.1306e-03 2.2344e+03 7.0095e+00]
[ 1.0011e+00 6.0150e-04 2.2430e+03 7.0011e+00]
[ 9.9902e-01 2.8231e-04 2.2570e+03 6.9856e+00]
[ 1.0004e+00 -1.8229e-03 2.2656e+03 7.0057e+00]]
>>>
>>> # All errors, frequencies in Hz
>>> estimator.get_errors()
[[0.0013 0.0013 0.0019 0.0121]
[0.0014 0.0014 0.002 0.0124]
[0.0014 0.0014 0.002 0.0125]
[0.0013 0.0013 0.0019 0.012 ]
[0.0012 0.0012 0.0018 0.0114]
[0.0036 0.0036 0.0034 0.0212]
[0.0036 0.0036 0.0034 0.0213]
[0.0012 0.0012 0.0018 0.0114]
[0.0013 0.0013 0.0019 0.0116]
[0.0013 0.0013 0.0019 0.0118]
[0.0013 0.0013 0.0019 0.0118]
[0.0013 0.0013 0.0018 0.0116]]
>>>
>>> # Params for first region, frequencies in ppm
>>> estimator.get_params(indices=[0], funit="ppm")
[[ 1.0003e+00 1.1306e-03 4.4688e+00 7.0095e+00]
[ 1.0011e+00 6.0150e-04 4.4860e+00 7.0011e+00]
[ 9.9902e-01 2.8231e-04 4.5140e+00 6.9856e+00]
[ 1.0004e+00 -1.8229e-03 4.5312e+00 7.0057e+00]]
>>>
>>> # Params for second and third regions, split up
>>> estimator.get_params(indices=[1, 2], merge=False, funit="ppm")
[array([[ 1.0022e+00, 7.1936e-04, 3.8772e+00, 7.0109e+00],
[ 9.9470e-01, -7.4609e-04, 3.9176e+00, 6.9866e+00],
[ 1.0080e+00, -1.0112e-03, 3.9224e+00, 7.0448e+00],
[ 1.0009e+00, -7.1398e-04, 3.9628e+00, 7.0131e+00]]),
array([[1.0018e+00, 1.5921e-03, 3.6712e+00, 7.0187e+00],
[1.0003e+00, 2.4881e-03, 3.6884e+00, 6.9968e+00],
[1.0024e+00, 1.5817e-03, 3.7116e+00, 7.0281e+00],
[1.0008e+00, 9.1591e-04, 3.7288e+00, 7.0007e+00]])]
Writing result tables¶
Tables of parameters can be saved to .txt and .pdf formats. using
write_result(). For PDF generation, you will
need a working LaTeX installation. See the installation instructions.
>>> for fmt in ("txt", "pdf"):
... estimator.write_result(
... path="tutorial_1d",
... fmt=fmt,
... description="Simulated 2,3-Dibromopropanoic acid signal.",
... )
...
Saved file tutorial_1d.txt.
Saved file tutorial_1d.tex.
Saved file tutorial_1d.pdf.
You can view and customise the corresponding TeX file at tutorial_1d.tex.
tutorial_1d.txt: Text file.tutorial_1d.pdf: PDF file.tutorial_1d.tex: TeX file used to generate the PDF
Creating result plots¶
Figures giving an overview of the estimation result can be generated using
plot_result().
>>> for (txt, indices) in zip(("complete", "index_1"), (None, [1])):
... fig, ax = estimator.plot_result(
... indices=indices,
... figure_size=(4.5, 3.),
... region_unit="ppm",
... axes_left=0.03,
... axes_right=0.97,
... axes_top=0.98,
... axes_bottom=0.09,
... )
... fig.savefig(f"tutorial_1d_{txt}_fig.pdf")
...
tutorial_1d_complete_fig.pdf: result for all regions consideredtutorial_1d_index_1_fig.pdf: result for 2nd estimated region only (index 1)
Saving the estimator¶
The estimator object itself can be saved and reloaded for future use with the
to_pickle() and
from_pickle() methods, respectively:
>>> estimator.to_pickle("tutorial_1d")
Saved file tutorial_1d.pkl.
>>> # Load the estimator and assign to the `estimator_cp` variable
>>> estimator_cp = ne.Estimator1D.from_pickle("tutorial_1d")
Saving a logfile¶
A logfile listing all the methods called on the estimator can be created using
save_log():
>>> estimator.save_log("tutorial_1d")
Saved file tutorial_1d.log.
The full script¶
Everything covered here is incorporated into the following Python script:
estimator1d_tutorial.py